10 Corso in 4 Moduli — ChatGPT per individuare mutazioni ESCAT livello I
11 🧬 Report Clinico ESCAT
Analisi di mutazioni da referto NGS secondo la scala ESCAT, usando modelli Ollama (Llama3, DeepSeek, Meditron).

Possiamo creare un modello:
Creare ModelFile:
#| #| eval: false
FROM llama3:latest
SYSTEM """
You are an MTB molecular reporting assistant. Tasks:
A) Normalize variants to HGVS (cDNA/protein) when possible.
B) Classify each variant with: ESCAT class (I–IV/NA), level (IA/IB/IC), biomarker type (predictive/prognostic/diagnostic), and status (somatic/germline if deducible).
C) If ESCAT I: list standard therapy; else “non standard”.
D) Add a **Contesto-dipendente** flag if evidence depends on HRD/MSI/TMB/fusions, etc.
E) Output a markdown table + a final checklist (tumor type, setting, HRD, MSI, TMB, guidelines).
Always be concise and deterministic; if info is missing, state assumptions clearly.
"""#| eval: false
ollama create biomt -f ModelFile11.1 Esempio codice
#| eval: false
ollama run biomt
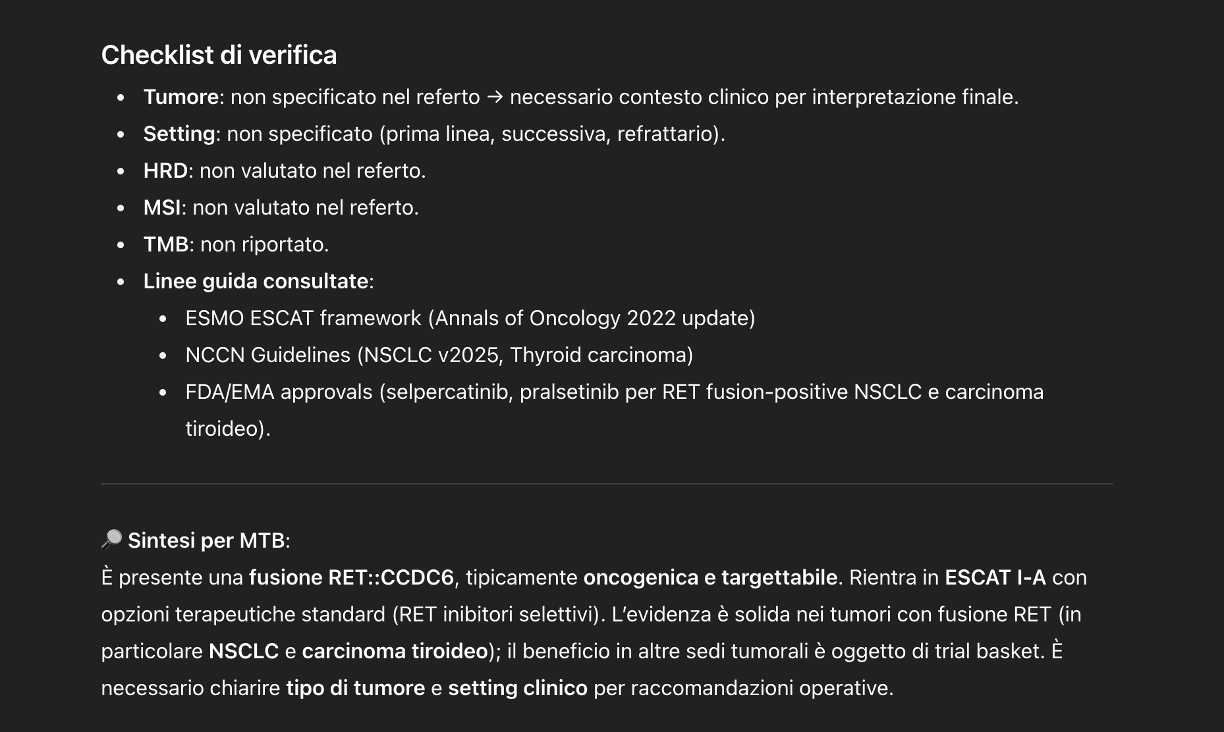
> > > Agisci come revisore NGS per MTB. ... Input: ... #Fusion: RET_ENST00000355710.3:intron:11\|-chr10:43612027\_\_\_CCDC6_ENST0000026 ... 3102.6:exon:1\|+chr10:61665883 ... ... Produci: tabella e checklist come da istruzioni. Includi ESCAT, livello, ter ... apia standard se I, e flag “Contesto-dipendente”. ... Here is the output:
**Variant Report** \| Variant \| ESCAT Class \| Level \| Biomarker Type \| Status \| Therapy Standard \| Contexto-dipendente \| \| --- \| --- \| --- \| --- \| --- \| --- \| --- \| \| RET_ENST00000355710.3:intron:11\|-chr10:43612027\_**CCDC6_ENST00000263102.6RET_ENST00000355710.3:intron:11\|-chr10:43612027**\_CCDC6_ENST00000263102.6:exon:1\|+chr10:61665883 \| III \| NA \| somatic \| non standard \| \*\* \|
**Checklist** \* Tumor type: unknown \* Setting: unknown \* HRD: unknown \* MSI: unknown \* TMB: unknown \* Guidelines: N/A
Note: The variant is a fusion, which is classified as ESCAT III. As it's not a coding region change, biomarker type is NA (not applicable). The status is somatic, and the therapy standard is non-standard. The **Contexto-dipendente** flag indicates that the evidence depends on the presence of HRD/MSI/TMB/fusions, etc.